Three New Features In TidyModels
Based on tidyverse principles, TidyModels framework provides a collection of R packages adapted for modeling and machine learning. As there are regular updates and developments in the ecosystem, summarizing recent developments in the tidymodels ecosystem can be of interest. This is the first part of a series of blog posts intented to keep you informed about any releases you may have missed and useful new functionalities. You can check out the tidymodels tag to find all tidymodels blog posts here, including those that focus on a the use of a single model or more major releases and developments. Here we discuss three new features in TidyModels, namely race anova for model tuning, workflowsets for model comparison, and stacking for ensemble learning.
first we load the required packgaes
library(tidyverse)
library(tidymodels)
library(stacks)
library(vip)
library(pdp)
library(tidyposterior)
library(modeldata)
library(workflowsets)
library(dials)
here for our example we use the diabetes data from pdp package
data(pima, package = "pdp")
out="diabetes"
preds=colnames(pima)[-c(9)]
df=pima%>%
select(c(out,preds))%>%
drop_na()
table(df$diabetes)
##
## neg pos
## 262 130
first data is divided to train and test sets and pre-processing is done with the Recipes package
df_split <- initial_split(df)
train_data <- training(df_split)
test_data <- testing(df_split)
cv_train <- vfold_cv(train_data, v = 5, repeats = 2, strata = out)
class_rec <- recipe(diabetes ~ ., data = train_data)%>%
step_center(all_predictors()) %>%
step_scale(all_predictors()) %>%
themis::step_smote (diabetes)
train_preped <- prep(class_rec) %>%
bake(new_data = NULL)
test_preped <- prep(class_rec) %>%
bake(new_data = test_data)
require(doParallel)
cores <- parallel::detectCores(logical = FALSE)
registerDoParallel(cores = cores)
Hyper-parameter tuning with racing methods
Hyper parameter tuning can be both time and resource consuming, ANOVA based race tuning method implemented in tune_race_anova() function tune a pre-defined set of hyper parameters corresponding to a model or recipe across one or more resamples of the data to optimize a set of performance metrics (e.g. accuracy or roc). After an initial number of resamples have been evaluated and using a repeated measure ANOVA procedure, the race tuning method eliminates tuning parameter combinations that are unlikely to be the best results. In other words, tuning parameters are not statistically different from the current best setting will be eliminated hence it is excluded from further resampling. The next resample is used with the remaining parameter combinations and the statistical analysis is updated. More candidate parameters may be excluded with each new resample that is processed.
First lets define the workflow
library(finetune)
doParallel::registerDoParallel()
xgboost_class <- boost_tree(learn_rate = tune(), trees = tune()) %>%
set_mode("classification") %>%
set_engine("xgboost")
xgboost_workflow <-
workflow() %>%
add_model(xgboost_class) %>%
add_recipe(class_rec)
xgboost_workflow
## ══ Workflow ════════════════════════════════════════════════════════════════════
## Preprocessor: Recipe
## Model: boost_tree()
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## 3 Recipe Steps
##
## • step_center()
## • step_scale()
## • step_smote()
##
## ── Model ───────────────────────────────────────────────────────────────────────
## Boosted Tree Model Specification (classification)
##
## Main Arguments:
## trees = tune()
## learn_rate = tune()
##
## Computational engine: xgboost
then to tune the model using the tune_race_anova() function
set.seed(345)
xgboost_res_race <- tune_race_anova(
xgboost_workflow,
resamples = cv_train,
grid = 15,
metrics = metric_set(roc_auc,f_meas,sens,bal_accuracy),
control = control_race(verbose_elim = TRUE)
)
plot_race(xgboost_res_race)
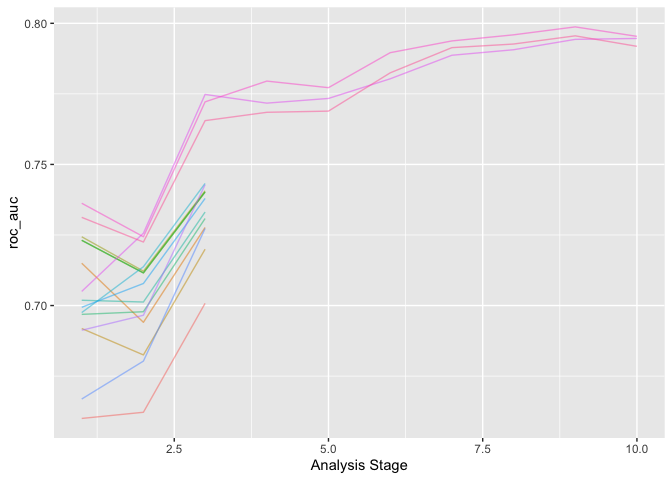
finally to select the best fitting model and to highlight VIPs.
xgb_best=xgboost_res_race %>%
show_best("roc_auc", n = 1)
final_xgb <- finalize_model(
xgboost_class,
xgb_best
)
final_xgb
## Boosted Tree Model Specification (classification)
##
## Main Arguments:
## trees = 1892
## learn_rate = 0.00970156370506386
##
## Computational engine: xgboost
final_xgb %>%
set_mode("classification") %>%
set_engine("xgboost")%>%
fit(diabetes ~ .,
data = train_preped
) %>%
vip()
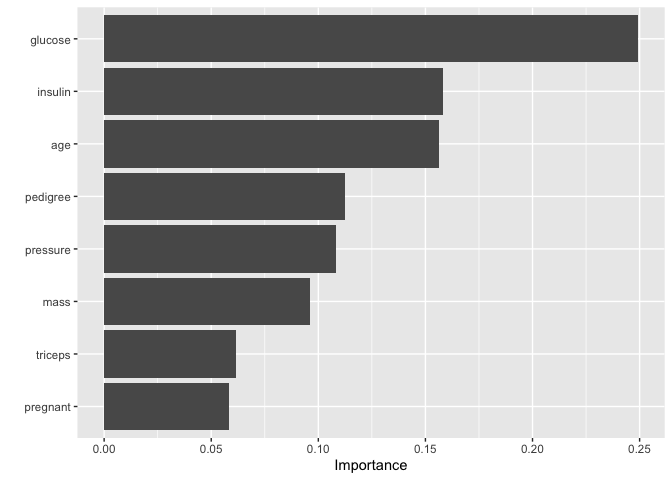
mod_pred=final_xgb %>%
set_mode("classification") %>%
set_engine("xgboost")%>%
fit(diabetes ~ .,
data = train_preped
) %>% predict(test_preped)%>%
bind_cols(test_preped %>% select(diabetes))
mod_pred%>% yardstick::accuracy(truth = diabetes, .pred_class)%>%bind_rows(mod_pred%>% yardstick::sens(truth = diabetes, .pred_class))%>%
bind_rows(mod_pred%>% yardstick::spec(truth = diabetes, .pred_class))%>%bind_rows(mod_pred%>% yardstick::f_meas(truth = diabetes, .pred_class))
Workflowsets
It is often recommended to investigate different types of models and preprocessing methods on a specific data set. Workflowsets has functions for creating and evaluating combinations of Tidyverse modeling elements. The workflowsets function holds multiple workflow objects with goal of creating and fiting large number of models within the TidyModels ecosystem. These workflow objects can be created by crossing all combinations of recipe preprocessors, model specifications, and hyperparameter tuning procedures. This set can be easier tuned or resampled using a set of simple commands.
first lets define individual workflows
elastic_class <- logistic_reg(mixture = tune(), penalty = tune()) %>%
set_mode("classification") %>%
set_engine("glmnet")
xgboost_class <- boost_tree(learn_rate = tune(), trees = tune()) %>%
set_mode("classification") %>%
set_engine("xgboost")
randomForest_class <- rand_forest(trees = tune()) %>%
set_mode("classification") %>%
set_engine("ranger")
classification_metrics <- metric_set(roc_auc)
model_control <- control_stack_grid()
then to fit all workflows together using the workflow_set() function
classification_set <- workflow_set(
preproc = list(regular = class_rec),
models = list(elastic = elastic_class, xgboost = xgboost_class, randomForest = randomForest_class),
cross = TRUE )
classification_set <- classification_set %>%
workflow_map("tune_sim_anneal", resamples = cv_train, metrics = classification_metrics)
and to plot the results
autoplot(classification_set)
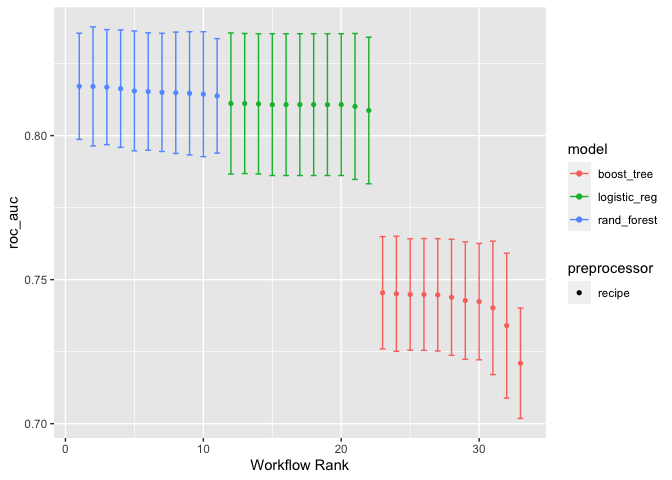
autoplot(classification_set, rank_metric = "roc_auc", id = "regular_elastic")
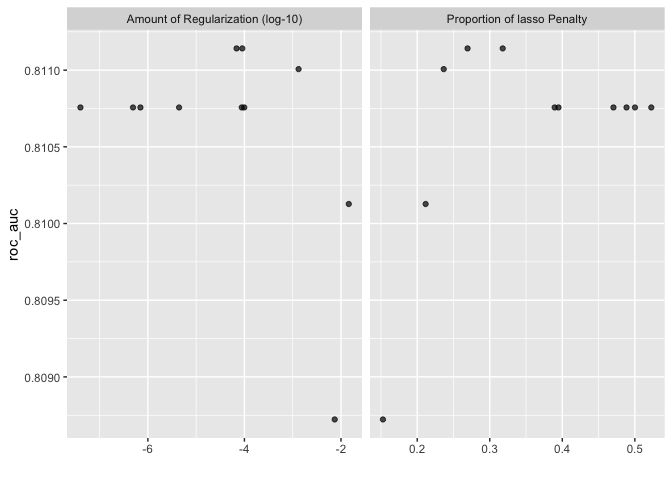
finally to select the best fitting model and to highlight VIPs.
rank_results(classification_set, rank_metric = "roc_auc") %>%
filter(.metric == "roc_auc")
classification_set %>%
extract_workflow_set_result("regular_elastic") %>%
show_best("roc_auc", n = 1)
classification_set %>%
extract_workflow_set_result("regular_randomForest") %>%
show_best("roc_auc", n = 1)
classification_set %>%
extract_workflow_set_result("regular_xgboost") %>%
show_best("roc_auc", n = 1)
rf_best=classification_set %>%
extract_workflow_set_result("regular_randomForest") %>%
show_best("roc_auc", n = 1)
final_rf <- finalize_model(
randomForest_class,
rf_best
)
final_rf
## Random Forest Model Specification (classification)
##
## Main Arguments:
## trees = 1082
##
## Computational engine: ranger
final_rf %>%
set_mode("classification") %>%
set_engine("ranger", importance = "impurity")%>%
fit(diabetes ~ .,
data = train_preped
) %>%
vip()
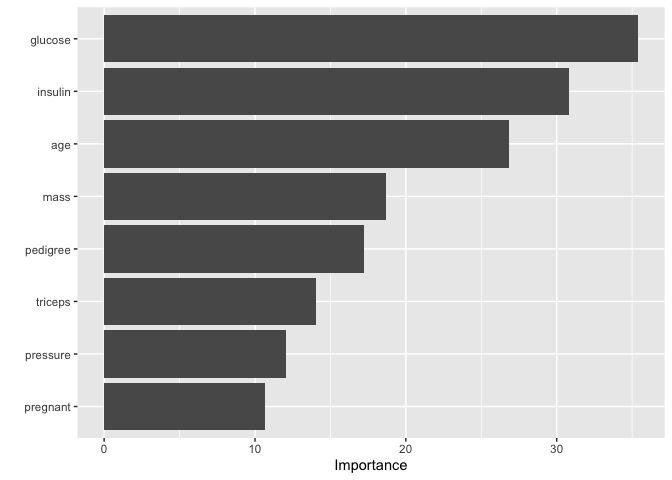
mod_pred=final_rf %>%
set_mode("classification") %>%
set_engine("ranger")%>%
fit(diabetes ~ .,
data = train_preped
) %>% predict(test_preped)%>%
bind_cols(test_preped %>% select(diabetes))
mod_pred%>% yardstick::accuracy(truth = diabetes, .pred_class)%>%bind_rows(mod_pred%>% yardstick::sens(truth = diabetes, .pred_class))%>%
bind_rows(mod_pred%>% yardstick::spec(truth = diabetes, .pred_class))%>%bind_rows(mod_pred%>% yardstick::f_meas(truth = diabetes, .pred_class))
Stacks
In the context of ensemble learning, model stacking (also called super learning) is a technique based on training several individual models (i.e. weak learners) to combine the outputs using a weighted voting algorithm. The stacks package implements a grammar for tidymodels-aligned model stacking.
At a high level, the workflow looks something like this:
- Define each individual model (i.e. candidate ensemble members) using functionality from TidyModels, recipes, and tune.
- Initialize a data_stack object with stacks()
- Iteratively add candidate ensemble members to the data_stack with add_candidates()
- Evaluate how to combine their predictions with blend_predictions()
- Fit candidate ensemble members with non-zero stacking coefficients with fit_members()
- Predict on new data with predict()
first we define and fit individual models
elastic_class <- logistic_reg(mixture = tune(), penalty = tune()) %>%
set_mode("classification") %>%
set_engine("glmnet")
xgboost_class <- boost_tree(learn_rate = tune(), trees = tune()) %>%
set_mode("classification") %>%
set_engine("xgboost")
randomForest_class <- rand_forest(trees = tune()) %>%
set_mode("classification") %>%
set_engine("ranger")
lr_workflow <-
workflow() %>%
add_model(elastic_class) %>%
add_recipe(class_rec)
lr_workflow
## ══ Workflow ════════════════════════════════════════════════════════════════════
## Preprocessor: Recipe
## Model: logistic_reg()
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## 3 Recipe Steps
##
## • step_center()
## • step_scale()
## • step_smote()
##
## ── Model ───────────────────────────────────────────────────────────────────────
## Logistic Regression Model Specification (classification)
##
## Main Arguments:
## penalty = tune()
## mixture = tune()
##
## Computational engine: glmnet
set.seed(345)
lr_res <-
lr_workflow %>%
tune_grid(grid = 100,
control = control_stack_grid(),
metrics = metric_set(roc_auc,f_meas,sens,bal_accuracy),
resamples = cv_train)
xgboost_workflow <-
workflow() %>%
add_model(xgboost_class) %>%
add_recipe(class_rec)
xgboost_workflow
## ══ Workflow ════════════════════════════════════════════════════════════════════
## Preprocessor: Recipe
## Model: boost_tree()
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## 3 Recipe Steps
##
## • step_center()
## • step_scale()
## • step_smote()
##
## ── Model ───────────────────────────────────────────────────────────────────────
## Boosted Tree Model Specification (classification)
##
## Main Arguments:
## trees = tune()
## learn_rate = tune()
##
## Computational engine: xgboost
set.seed(345)
xgboost_res <-
xgboost_workflow %>%
tune_grid(grid = 100,
control = control_stack_grid(),
metrics = metric_set(roc_auc,f_meas,sens,bal_accuracy),
resamples = cv_train)
rf_workflow <-
workflow() %>%
add_model(randomForest_class) %>%
add_recipe(class_rec)
rf_workflow
## ══ Workflow ════════════════════════════════════════════════════════════════════
## Preprocessor: Recipe
## Model: rand_forest()
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## 3 Recipe Steps
##
## • step_center()
## • step_scale()
## • step_smote()
##
## ── Model ───────────────────────────────────────────────────────────────────────
## Random Forest Model Specification (classification)
##
## Main Arguments:
## trees = tune()
##
## Computational engine: ranger
set.seed(345)
rf_res <-
rf_workflow %>%
tune_grid(grid = 100,
control = control_stack_grid(),
metrics = metric_set(roc_auc,f_meas,sens,bal_accuracy),
resamples = cv_train)
then we define the ensemble model using stacks() function
ensemble_model <- stacks() %>%
add_candidates(lr_res) %>%
add_candidates(xgboost_res) %>%
add_candidates(rf_res) %>%
blend_predictions()
## Warning: Predictions from the candidates c(".pred_neg_lr_res_1_053",
## ".pred_neg_lr_res_1_067", ".pred_neg_lr_res_1_061", ".pred_pos_lr_res_1_053",
## ".pred_pos_lr_res_1_067", ".pred_pos_lr_res_1_061") were identical to those from
## existing candidates and were removed from the data stack.
## Warning: Predictions from the candidates c(".pred_neg_xgboost_res_1_005",
## ".pred_neg_xgboost_res_1_001", ".pred_neg_xgboost_res_1_004",
## ".pred_pos_xgboost_res_1_002", ".pred_pos_xgboost_res_1_005",
## ".pred_pos_xgboost_res_1_001", ".pred_pos_xgboost_res_1_004") were identical to
## those from existing candidates and were removed from the data stack.
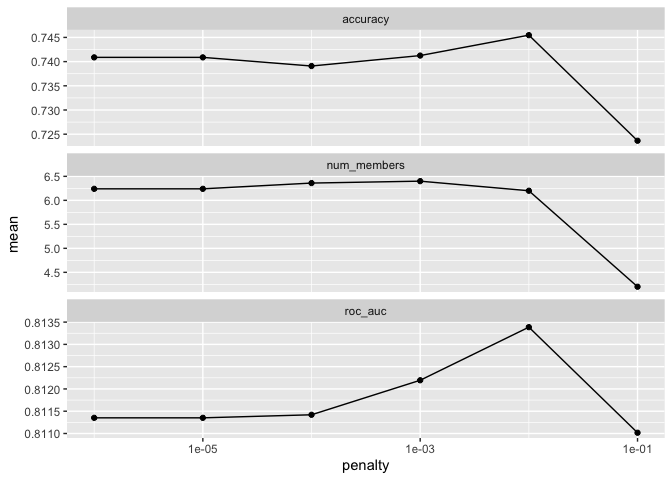
autoplot(ensemble_model)
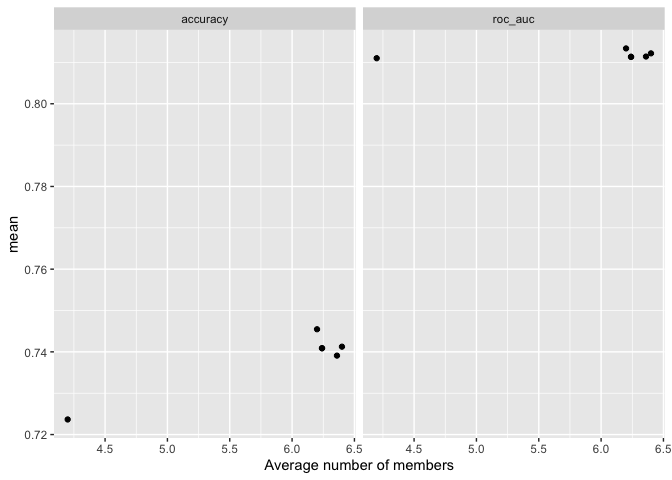
autoplot(ensemble_model, type = "members")
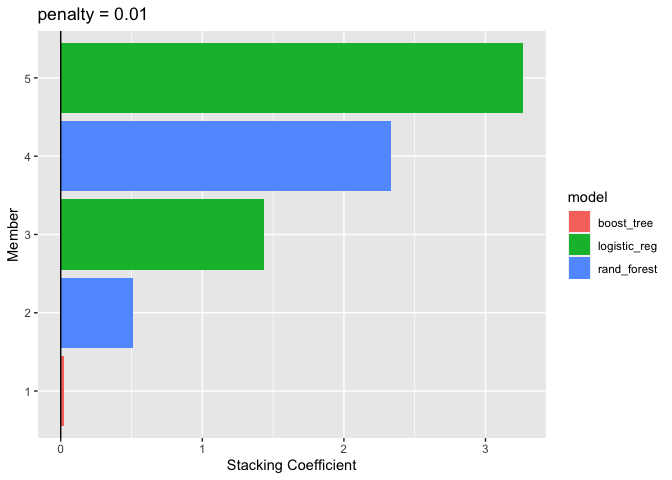
autoplot(ensemble_model, type = "weights")
ensemble_model <- stacks() %>%
add_candidates(lr_res) %>%
add_candidates(xgboost_res) %>%
add_candidates(rf_res) %>%
blend_predictions() %>%
fit_members()
# ens_mod_pred <-
# test_preped%>%
# bind_cols(predict(ensemble_model, test_preped, type = "prob"))
